Water structure and dynamics at (bio)-interfaces and in confinement
Water plays a major role in biosystems, greatly contributing to determine their structure, stability, and function. A “special” role is played by the protein hydration water shells (Fig. 1a). These water molecules display physico-chemical properties that are apparently different from molecules in pure bulk water. For instance, they are prevented from crystallising just below 0 °C because the competition of their mutual interaction and their hydrogen bonding to the protein makes it difficult for them to arrange into the typical tetrahedral ice structure. Remarkably, proteins’ denaturation restores water ability to crystallise (Fig. 1b). In this context, we shown for the first time, by means of high-resolution elastic neutron scattering, a de-coupling between protein and its hydration water dynamics (Fig. 2). More in general, we are interest in the structure and dynamical behaviour of water molecules confined in both biological and non biological matrixes. These include water solutions of ionic liquids in which, depending on concentrations, nano domains of either water or ionic liquids can form (Fig. 3) up to the mesoscopic phase separations of thermoresponsive ionic liquids (Fig. 4). We are also interested in the role of water in biophysical systems including, for example, in the partitioning of ionic liquids between lipid and aqueous phases in hydrated lipid bilayers (Fig. 5) and in protein amyloidogenesis (Fig. 6). Neutron scattering and computer simulations are the two major approaches used in this research line.
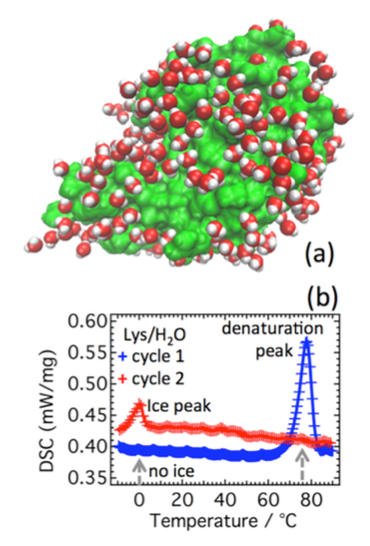
Fig. 1 - Model protein lysozyme and its hydration water. (a) Crystal structure of human lysozyme (green) together with ∼280 hydration water molecules obtained by X-ray diffraction at 1.8 Å of spatial resolution. (b) Differential scan calorimetric heating profiles before (blue) and after (red) thermal denaturation (blue peak). The hydration water due to the interaction with the protein is not able to form ice, but after the thermal denaturation, due to a different state of the protein, part of the hydration water molecules recover their ability to form ice (red peak). Taken from our J Phys Chem Lett 2017
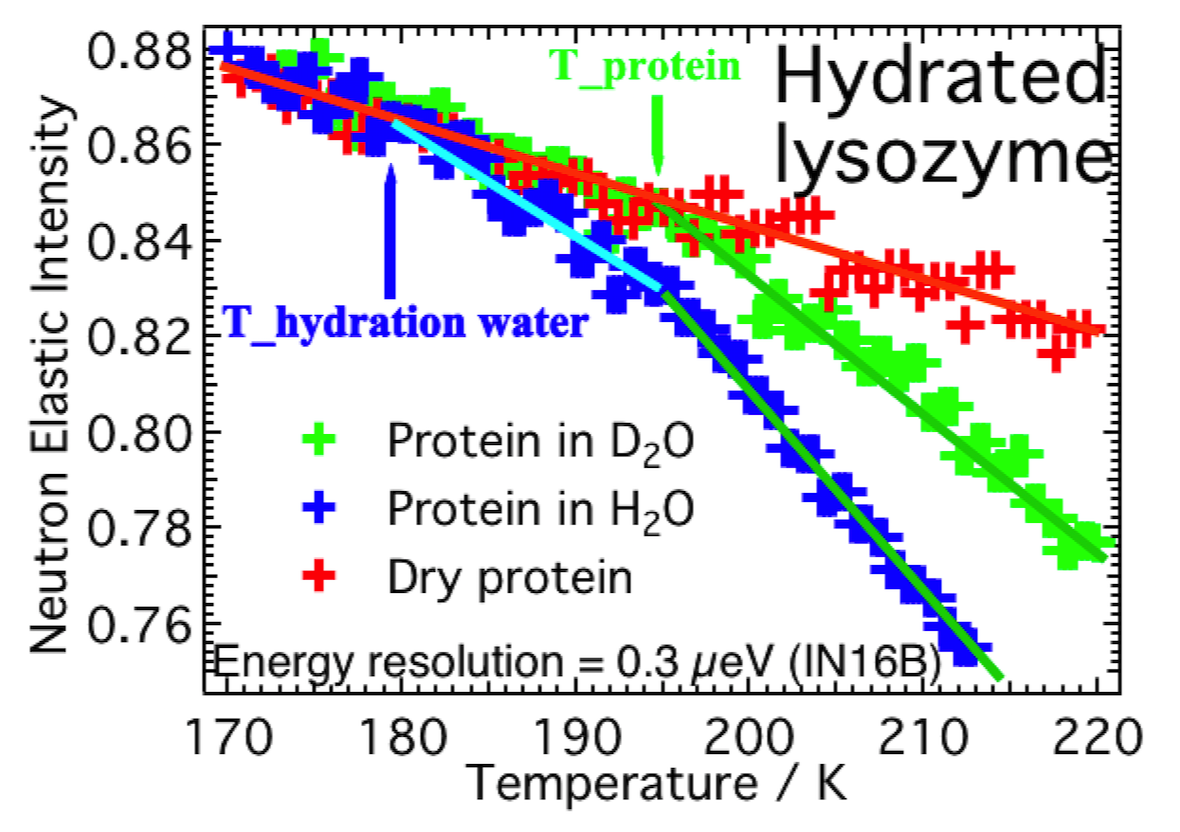
Fig. 2 - High-resolution elastic neutron scattering data showing the decoupling between the dynamics of protein and of its hydration water. Total elastic neutron-scattered intensity versus temperature for D2O hydrated lysozyme (green), H2O hydrated lysozyme (blue), and dry lysozyme (red). The D2O signal from a protein hydrated in D2O is negligible, and relaxations in the elastic spectrum can be related to the protein itself, which also contains a high number of hydrogen atoms. In this way it is possible to probe the relaxation dynamics of protein alone, yet hydrated. In the case of a protein hydrated in H2O, the contributions of water and protein have very similar weights in the measured signal (i.e., the densities of hydrogen atoms in the protein and in hydration water are nearly equivalent). As a result, by measuring the protein hydrated in H2O, relaxations in the measured elastic spectrum arise from relaxations of either the protein or its hydration water. From this set of data it was possible to determine that the protein starts to relax at T_p = 195 K, decoupled from the transition in the dynamics of hydration water, which starts to relax 16 K lower, that is, T_hw = 179 K. Taken from our J Phys Chem Lett 2017
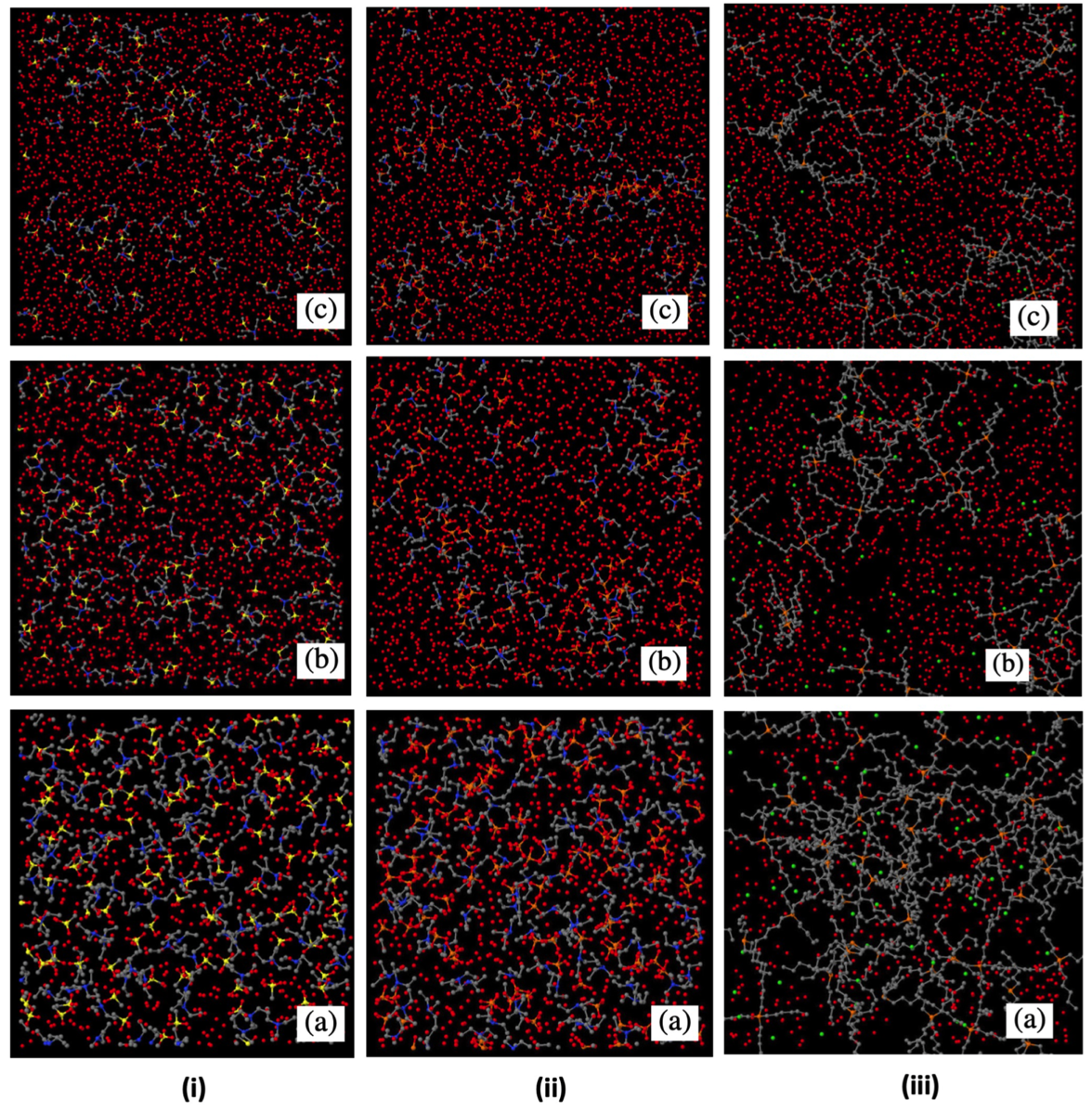
Fig. 3 - MD simulation snapshots for the (i) triethyl ammonium mesylate/water, (ii) triethyl ammonium dihydrogen phosphate/water and (iii) the phosphonium di-cation [DxC10][Cl]2/water systems for different concentration of water, i.e. number of water molecules (Nw) of: (a) Nw = 1075; (b) Nw = 4075; (c) Nw = 8075. Hydrogen atoms have been removed, hence water is represented by a single red dot. Nano domains can be observed by eye inspection. Taken from our Phys Chem Chem Phys 2021
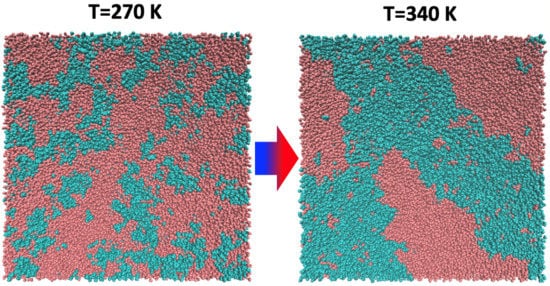
Fig. 4 - MD simulation snapshots showing a 50-50 wt% [P4444][DMBS]/water solution below and above the demixing temperature. [P4444] is an alkyl-phosphonium cation with 4 carbon atoms in each alkyl chain, [DMBS] stays for 2,4-dimethylbenzene sulfonate. Taken from our Molecules 2022
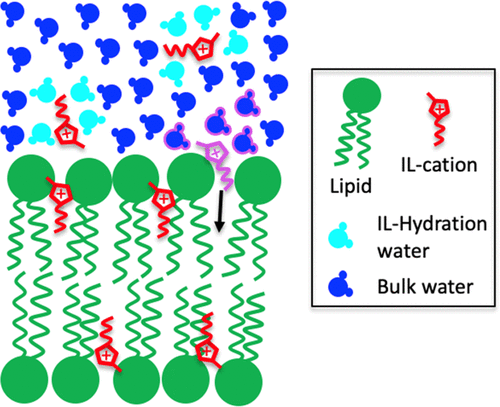
Fig. 5 - Sketch of the ionic liquids' hydration water entropy-driven absorption mechanism proposed to explain the observed increase in the number of ionic liquids cations absorbed in the lipid bilayer in its fluid phase with increasing temperature. When the purple-highlighted ionic liquid cation diffuses into the lipid bilayer, its pink-circled hydration water molecules get released into the bulk solvent. The entropy variation associated with the release of these water molecules is positive and could be the driving force governing the ionic liquid partitioning. Taken from our J Phys Chem B 2022
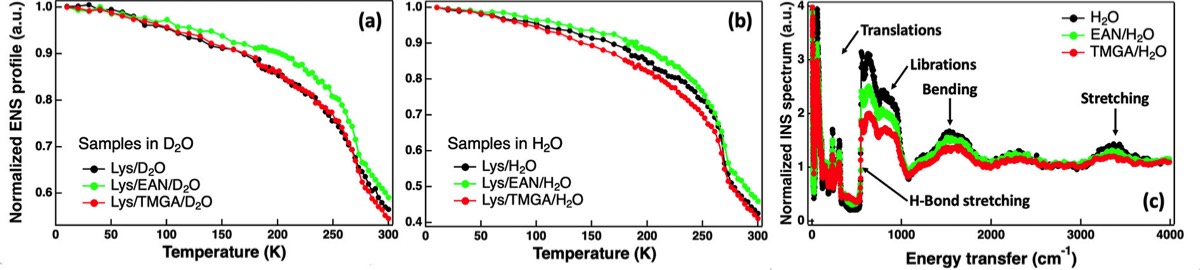
Fig. 6 - (a) Elastic neutron scattering intensity as a function of temperature for lysozyme hydrated in heavy water (black) and heavy water solutions of ethyl ammonium nitrate (EAN) in green and tetramethyl guanidinium acetate (TMGA) in red at a molar ratio of 2 ionic liquids per protein. (b) As in panel a but with water instead of heavy water. These results show that TMGA interacts with the protein hydration layer only, making the relaxation dynamics of these water molecules faster. EAN interacts directly with the protein instead, slowing down its relaxation dynamics. The stronger interaction between TMGA and water was confirmed by inelastic neutron scattering. (c) Inelastic neutron scattering spectra of pure water (black) and water solutions of EAN (green) and TMGA (red) at 0.5 M. Taken from our J Phys Chem Lett 2022
Selected publications:
Controlling Amyloid Fibril Properties Via Ionic Liquids: The Representative Case of Ethylammonium Nitrate and Tetramethylguanidinium Acetate on the Amyloidogenesis of Lysozyme
V. Pillai, P. Kumari, S. Kolagatla, V.G. Sakai, S. Rudić, B. Rodriguez, M. Rubini, K. Tych, A. Benedetto, Journal of Physical Chemistry Letters, 13, 7058, (2022)
>>>> LINK TO THE PAPER <<<<
>>>> Featuring on the JOURNAL COVER <<<<
V. Pillai, P. Kumari, S. Kolagatla, V.G. Sakai, S. Rudić, B. Rodriguez, M. Rubini, K. Tych, A. Benedetto, Journal of Physical Chemistry Letters, 13, 7058, (2022)
>>>> LINK TO THE PAPER <<<<
>>>> Featuring on the JOURNAL COVER <<<<
Absorption of the [bmim][Cl] Ionic Liquid in DMPC Lipid Bilayers Across Their Gel, Ripple, and Fluid Phases
A. Benedetto, E. Kelley, Journal of Physical Chemistry B, 126, 3309, (2022)
>>>> LINK TO THE PAPER <<<<
>>>> Featuring on the JOURNAL COVER <<<<
A. Benedetto, E. Kelley, Journal of Physical Chemistry B, 126, 3309, (2022)
>>>> LINK TO THE PAPER <<<<
>>>> Featuring on the JOURNAL COVER <<<<
Thermoresponsive Ionic Liquid/Water Mixtures: From Nanostructuring to Phase Separation
N.C. Forero-Martinez, R. Cortes-Huerto, A. Benedetto, P. Ballone, Molecules, 27, 1647, (2022)
>>>> LINK TO THE PAPER <<<<
N.C. Forero-Martinez, R. Cortes-Huerto, A. Benedetto, P. Ballone, Molecules, 27, 1647, (2022)
>>>> LINK TO THE PAPER <<<<
The transition from salt-in-water to water-in-salt nanostructures in water solutions of organic ionic liquids relevant for biological applications
P. Kumari, V. Pillai, D. Gobbo, P. Ballone, A. Benedetto, Physical Chemistry Chemical Physics, 125, 7241, (2021)
>>>> LINK TO THE PAPER <<<<
P. Kumari, V. Pillai, D. Gobbo, P. Ballone, A. Benedetto, Physical Chemistry Chemical Physics, 125, 7241, (2021)
>>>> LINK TO THE PAPER <<<<
Low-Temperature Decoupling of Water and Protein Dynamics Measured by Neutron Scattering
A. Benedetto, The Journal of Physical Chemistry Letters, 8, 4883, (2017)
>>>> LINK TO THE PAPER <<<<
A. Benedetto, The Journal of Physical Chemistry Letters, 8, 4883, (2017)
>>>> LINK TO THE PAPER <<<<
Hydrogen-Bond Dynamics at the Bio-Water Interface in Hydrated Proteins: a Molecular Dynamics Study
P. Nandi, N. English, Z. Futera, and A. Benedetto, Physical Chemistry Chemical Physics, 19, 318, (2017)
>>>> LINK TO THE PAPER <<<<
P. Nandi, N. English, Z. Futera, and A. Benedetto, Physical Chemistry Chemical Physics, 19, 318, (2017)
>>>> LINK TO THE PAPER <<<<
Puzzle of Protein Dynamical Transition
S. Magazù, F. Migliardo, A. Benedetto, Journal of Physical Chemistry B, 115, 7736, (2011)
>>>> LINK TO THE PAPER <<<<
S. Magazù, F. Migliardo, A. Benedetto, Journal of Physical Chemistry B, 115, 7736, (2011)
>>>> LINK TO THE PAPER <<<<
Mean Square Displacements from Elastic Incoherent Neutron Scattering Evaluated by Spectrometers Working with Different Energy Resolution on Dry and Hydrated (H2O and D2O) Lysozyme
S. Magazù, F. Migliardo, A. Benedetto, Journal of Physical Chemistry B, 114, 9268, (2010)
>>>> LINK TO THE PAPER <<<<
S. Magazù, F. Migliardo, A. Benedetto, Journal of Physical Chemistry B, 114, 9268, (2010)
>>>> LINK TO THE PAPER <<<<